Artificial intelligence has won its first Nobel Prizes – including for groundbreaking life science tools – and generative AI is ever-more powerful and prevalent. SIB is catalysing, enabling, and harnessing these and other transformative advances in AI.
SIB expertise and data pivotal to Nobel-winning AI model
AlphaFold was recognized in the 2024 Nobel Prize in Chemistry for its ability to predict 3D protein structures from their amino acid sequence. The model was built on decades of bioinformatics expertise – including three open resources and initiatives developed and co-developed by SIB scientists:
- The AI model learned to identify relationships between amino acid sequences and 3D structures by analysing hundreds of millions of high-quality protein sequences in UniProt. Expert annotations on protein structure also helped AlphaFold’s developers to understand and debug model performance.
- Its impressive accuracy was first demonstrated by CASP – a global competition held every two years that tests the accuracy of computational methods against experimentally determined, unpublished, and very challenging protein structures. SIB Group Leader Torsten Schwede has been a member of the CASP organizing committee since 2011.
- AlphaFold’s applicability to all human proteins was subsequently confirmed using data from CAMEO, which performs the same accuracy test as CASP but on a larger set of proteins published weekly.
See how SIB scientists mapped the protein universe from AlphaFold data
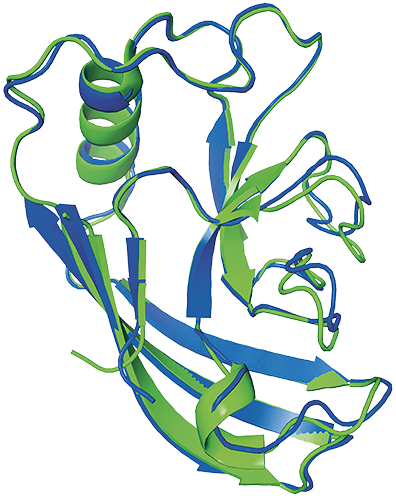
Protein structure predicted by AlphaFold (blue) and experimentally (green)
Providing gold-standard training data through curated databases
SIB’s curated databases provide highly reliable data and knowledge from which AI models can learn to recognize patterns and make relevant predictions. One of these, UniProt, was crucial in training AlphaFold (see box). Many others are available for, and being used in, AI applications to tackle complex challenges across the life sciences.
SIB expertise and data pivotal to Nobel-winning AI model
AlphaFold was recognized in the 2024 Nobel Prize in Chemistry for its ability to predict 3D protein structures from their amino acid sequence. The model was built on decades of bioinformatics expertise – including three open resources and initiatives developed and co-developed by SIB scientists:
- The AI model learned to identify relationships between amino acid sequences and 3D structures by analysing hundreds of millions of high-quality protein sequences in UniProt. Expert annotations on protein structure also helped AlphaFold’s developers to understand and debug model performance.
- Its impressive accuracy was first demonstrated by CASP – a global competition held every two years that tests the accuracy of computational methods against experimentally determined, unpublished, and very challenging protein structures. SIB Group Leader Torsten Schwede has been a member of the CASP organizing committee since 2011.
- AlphaFold’s applicability to all human proteins was subsequently confirmed using data from CAMEO, which performs the same accuracy test as CASP but on a larger set of proteins published weekly.
See how SIB scientists mapped the protein universe from AlphaFold data

Protein structure predicted by AlphaFold (blue) and experimentally (green)
- Transforming data into AI-ready knowledge
Our databases are open, FAIR, and machine-readable, achieved through harmonized data, rich metadata, and rigorous quality control.
Expertly curated databases additionally include relevant and continuously updated information from the scientific literature and other sources. Our biocurators annotate protein sequences in UniProt, for example, with knowledge on the proteins’ structure, function, and more.
This explicit encoding of complex information in machine-readable formats is essential for AI advances in the scientific domain. - A wealth of freely available training data
SIB co-develops open, curated databases spanning biomolecules and processes across the tree of life.
Examples include gene expression (Bgee), cell lines (Cellosaurus), carbohydrates (Glyco@Expasy), reactions (Rhea), protein interactions (STRING), lipids (SwissLipids), pathogen genomes (SPSP), orthologs (SwissOrthology), protein sequences and structures (UniProt, Swiss-Model), and gene functions (Gene Ontology). - Enabling training on sensitive data
We also develop ethical, secure methods for AI analyses of human data that cannot be openly shared. Federated analysis, for example, enables relevant data to be accessed in a secure environment without sharing any personal information.
SIB scientists are taking this approach in several initiatives, including the international iCARE4CVD initiative for personalized prevention and treatment of cardiovascular disease (read more) and BioMedIT, the trusted research environment of the Swiss Personalized Health Network (SPHN; read more).
Enabling actionable insights through knowledge representation
A single database is far more powerful when the knowledge it contains is linked to other databases. SIB scientists bridge data silos across fields and countries by developing and implementing systems to formally represent knowledge – such as standardized vocabularies to represent biological entities and their relationships (e.g., genes, proteins, metabolites, species, diseases).
This enables AI and researchers to effectively retrieve and process data from diverse sources, uncover connections between these data – and thus gain a holistic, integrated view of complex biological systems.
Examples include:
- developing AI-based discovery across the entire collection of European research outputs, through the European Open Science Cloud (EOSC) Data Commons project (read more);
- enabling AI-driven precision oncology by converting unstructured PDF guidelines for cancer care into structured, interoperable treatment procedures, and building an AI tool to analyse the procedures plus historical clinical data to predict the best treatment for individual patients, in the AI Tumor Board project with Swiss hospitals;
- fostering AI-assisted querying of curated plant knowledge by linking chemical data on plants to information such as species interactions and traits in the Digital Botanical Gardens Initiative.
Ensuring trustworthy outputs through benchmarking
How can AI developers – and the users of their models – be confident of AI predictions? And how can researchers know which AI tool is best for a particular task? SIB’s benchmarking expertise answered these questions for AlphaFold (see box above), and enables rigorous evaluation and fine-tuning of many other AI and computational analyses.
Our scientists:
- provide gold-standard reference datasets that serve as standardized inputs for comparing AI tools, as target outcomes against which predictions can be evaluated, and as resources for fine-tuning models for specific fields or assessing data quality before analyses;
- develop benchmarking software tools to assess AI performance against reference data or other models, highlight areas for improvement, and help researchers select the most suitable model for their needs.
Examples include tools and datasets for assessing protein-folding and protein-interaction predictions (CAMEO and CASP; see box above), proteome quality (SwissOrthology), and genome and metagenome quality (BUSCO and LEMMI); a dataset to fine-tune generative AI models (see below) for specialized data curation (EnzChemRED); and a system that ensures reproducible, neutral benchmarking of computational biology tools following FAIR (Findable, Accessible, Interoperable and Reusable) principles (Omnibenchmark).
Of generative AI and LLMs
Generative AI encompasses systems capable of creating new content, from text and images to videos, music and much more. Large Language Models (LLMs), a key type of generative AI, are trained on extensive text data, including genetic sequences or informatic code, to summarize, generate and predict new content. Models such as ChatGPT and BioBERT exemplify this, with ChatGPT excelling in generating text for chatbots and creative writing, while BioBERT focuses on (i.e., is pre-trained on) biomedical text. LLMs employ deep-learning techniques, particularly transformers, to analyse and understand language patterns from vast datasets, and to predict the next ‘word’ or sequence of words based on context.
Accelerating life science discoveries through generative AI
ChatGPT and a wealth of other generative AI models are not only disrupting our everyday lives, but also science. SIB is embracing this revolution across domains from clinical applications to biological knowledge generation.
Of generative AI and LLMs
Generative AI encompasses systems capable of creating new content, from text and images to videos, music and much more. Large Language Models (LLMs), a key type of generative AI, are trained on extensive text data, including genetic sequences or informatic code, to summarize, generate and predict new content. Models such as ChatGPT and BioBERT exemplify this, with ChatGPT excelling in generating text for chatbots and creative writing, while BioBERT focuses on (i.e., is pre-trained on) biomedical text. LLMs employ deep-learning techniques, particularly transformers, to analyse and understand language patterns from vast datasets, and to predict the next ‘word’ or sequence of words based on context.
Examples include:
- accelerating biological and bioinformatics discovery with ExpasyGPT – a customized generative AI tool integrated into Expasy, the Swiss bioinformatics resource portal, that allows researchers to retrieve and compile information from SIB databases more quickly and easily than ever (read more);
- fast generation of custom antibodies to fight disease through AntibodyGPT, which speeds up the traditionally slow process of discovering monoclonal antibodies by predicting antibody structures with desired properties;
- testing the ability of ChatGPT to answer medical questions in radiation therapy (read publication);
- deciphering the hidden role of RNA in cancer through expertise in Natural Language Processing;
- understanding how insects shed their skin by integrating species name data with sequence data from different public databases using generative AI methods into the MoultDB resource, serving as a reference for the field.
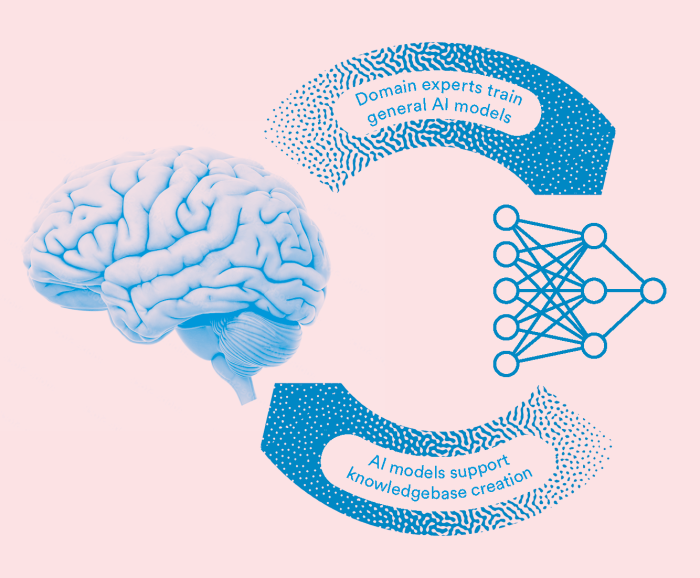
Generative AI and biocuration: a virtuous cycle
The interplay between the possibilities offered by AI, and LLMs in particular, and the importance of human expertise is well illustrated in the context of biocuration, where SIB is a recognized leader. Biocuration is the art of expertly extracting knowledge from the biological and biomedical literature to build an accurate, reliable and up-to-date encyclopedia serving science at large.
In-depth features on trustworthy AI
- The AI revolution continues – SIB Profile 2025
- Generative AI: new horizons – SIB Profile 2024
- Around machine learning – SIB Profile 2021