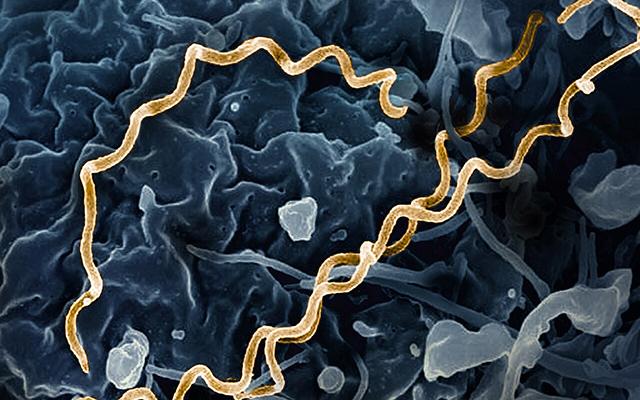
A bacterial genome found in the remains of an ancient hunter-gatherer shows that treponemal diseases were circulating far earlier in the Americas, and were more diverse, than previously known. Published in Science by an international team including SIB scientists at the University of Lausanne, the study represents the first recovery of Treponema pallidum DNA from healthy-looking bone. It also introduces a novel combination of bioinformatics methods for analysing ancient pathogen DNA – opening new avenues for understanding how infectious diseases emerge and spread.
Bioinformatics reveals ‘invisible’ pathogen in ancient human remains
Infections caused by T. pallidum include syphilis, yaws and bejel, which remain public health concerns today. Skeletal evidence from archaeological sites worldwide suggests these treponemal diseases have affected humans for millennia, yet their evolutionary origins have been hotly debated.
Paleogenomics can help answer such questions through the sequencing and analysis of pathogen DNA in ancient human remains. A key obstacle for T. pallidum, however, is the difficulty of recovering its DNA: the bacterium is scarce in bones, and only a small fraction of ancient skeletons shows signs of infection.
Now, for the first time, researchers have recovered T. pallidum DNA in healthy-looking bone from an individual buried around 5,500 years ago in what is now Colombia. Moreover, the international team identified the bacterium as a new subspecies. The novel combination of genomics analyses – carried out by SIB's Population Genetics Group, led by Anna-Sapfo Malaspinas at the University of Lausanne – push back the genetic record of T. pallidum by 3,000 years and suggest the pathogen's ancestors may have infected people as far back as the last Ice Age, 10-20,000 years ago. This significantly changes our understanding of treponemal diseases in the Americas and opens the door to improved estimates of disease prevalence throughout human history.
Different subspecies causing different diseases
The three present-day T. pallidum subspecies are nearly identical genetically, but each causes a distinct disease: T. pallidum subsp. pallidum causes syphilis and is sexually transmitted, while T. pallidum subsp. pertenue and T. pallidum subsp. endemicum, which spread primarily through direct skin contact, cause yaws and bejel respectively. A fourth treponemal disease, pinta, is caused by T. carateum, although no genome for this pathogen has yet been recovered.
The researchers believe the new subspecies they discovered likely spread person-to-person, but cannot rule out a zoonotic transmission from animals. It is also possible that it represents an ancient form of T. carateum. Further studies are needed to clarify these points.
T. pallidum ancestor may have infected people in the Ice Age
The SIB scientists and researchers at the University of California, Santa Cruz (UCSC), independently spotted the bacterial DNA while analysing sequencing data from the ancient hunter-gatherer. Despite comprising less than 0.002% of these data, the sheer volume – around 1.5 billion DNA fragments – allowed the researchers to reconstruct an ancient genome.
Different subspecies causing different diseases
The three present-day T. pallidum subspecies are nearly identical genetically, but each causes a distinct disease: T. pallidum subsp. pallidum causes syphilis and is sexually transmitted, while T. pallidum subsp. pertenue and T. pallidum subsp. endemicum, which spread primarily through direct skin contact, cause yaws and bejel respectively. A fourth treponemal disease, pinta, is caused by T. carateum, although no genome for this pathogen has yet been recovered.
The researchers believe the new subspecies they discovered likely spread person-to-person, but cannot rule out a zoonotic transmission from animals. It is also possible that it represents an ancient form of T. carateum. Further studies are needed to clarify these points.
Phylogenetic analysis revealed the bacterium is not only a new T. pallidum subspecies, but that it belongs to a previously unknown branch of the bacterium’s family tree. The SIB team confirmed this unexpected finding using multiple bioinformatics approaches, including methods not typically used in ancient pathogen studies. This included analysing shared genes across distantly related species using the OMA knowledge base of the SwissOrthology resource, developed by other SIB Groups, and mapping ancient and modern DNA with the Mapache pipeline developed by the Population Genetics group. Two other SIB Resources were also used: BEAST2 to calculate evolutionary timelines and UniProt’s expertly curated knowledgebase to examine protein structures.
The analysis also found all 59 virulence-associated genes known from today's strains in the new subspecies, suggesting their common ancestor may also have been adapted to human hosts. The researchers infer this ancestor lived approximately 13,700 years ago, though the true date could range from 7,000 to 20,500 years ago. Such diversity in the T. pallidum family – and such a long history of treponemal diseases in people, possibly as far back as the Ice Age – were unsuspected from previous ancient and modern genomic data.
New possibilities for understanding how pathogens emerge and spread
The study’s combination of advanced sequencing and novel genomics analyses could enable more widespread detection of infectious disease in other archaeological remains. This would help improve our understanding of how pathogens emerge, spread and persist within human populations – an essential first step to controlling and preventing future outbreaks and epidemics.
International collaboration with local community engagement
In addition to the SIB and UCSC researchers, the study brought together experts in paleopathology, anthropology, microbiology and archaeology from many other institutions across Switzerland, the USA, Colombia, Argentina, the Czech Republic and France. Recognizing the importance of the findings for Colombian medical and cultural history, the team consulted and shared their results with local scholars, students, and Indigenous and non-Indigenous communities before publishing the work.
See news from University of Lausanne (French)
See news from Southern Methodist University
Reference(s)
Bozzi D, Broomandkhoshbacht N, Delgado M et al. A 5,500-year-old Treponema pallidum genome from Sabana de Bogotá, Colombia. Science (2026).
Image: Electron micrograph image of modern T. pallidum, highlighted in gold. NIAID CC BY 2.0