An international study shows the unprecedented ability of the Pneumocystis fungus to outwit the human immune system. Philippe Hauser at the Lausanne University Hospital (CHUV) led the work, and the uniqueness of the molecular mechanisms involved was revealed based on bioinformatics analyses led by Marco Pagni at SIB. The findings are published in Nature Communications.
Microbial parasites that cause serious illnesses in humans, such as malaria or sleeping sickness, must evade the immune system to multiply in the human body. The laboratory of Philippe Hauser (CHUV) has been collaborating with SIB’s Marco Pagni and other key partners, to study the mechanisms used to achieve this by the Pneumocystis fungus, which causes lethal pneumonia in immunocompromised patients, those receiving organ transplants or those suffering from blood cancer, for example.
A new molecular mechanism of antigenic variation revealed
The study, published in the journal Nature Communications, reveals that this pathogen uses random shuffling of thousands of its genes occurring across its worldwide populations, to create a novel set of 80 genes in each strain. Within this set, a single gene is then expressed per individual cell of the fungus, to cover its surface with a 'coat' that the immune system cannot recognize and fight, because it has generally never encountered it before. This molecular mechanism of antigenic variation at cell level was hitherto unreported, and adds to those already known, for example in the trypanosome that causes sleeping sickness. Individual cells of the fungus then frequently change their 'coat' to avoid recognition by the immune system that develops during infection. Understanding these unique camouflage mechanisms, could lead to a new therapeutic strategy to combat the disease.
Building on 15 years of research
This discovery is the culmination of fifteen years' research on the pathogen, during which Philippe Hauser's team worked closely with Marco Pagni, Team Lead Computational Biology at SIB’s Vital-IT group, for the bioinformatics part. For the current study, SIB developed an essential computer procedure making it possible to differentiate the genes making up the set present in each strain of the fungus.
“Pneumocystis is a very difficult pathogen to work on. As it cannot be cultivated in vitro, we need to be able to say more with less, and this is where bioinformatics comes in. During our long research collaboration with Philippe Hauser’s lab, we started by sequencing its genome, then zoomed in on specific regions that are very complicated to assemble – doing all that from few precious clinical samples,” says Marco Pagni. “It’s thus fantastic to be today able to document a new mechanism explaining its ability to evade our immune system as a result.”
The study results from an international collaboration of researchers from the University of Lisbon, the CHU in Brest, the University of Cincinnati and the Institute of Biomedicine in Seville, as well as the Institute of Infectious Diseases at the University of Bern in Switzerland.
Reference(s)
Meier C S et al. Fungal antigenic variation using mosaicism and reassortment of subtelomeric genes’ repertoires, Nature Communications 2023.
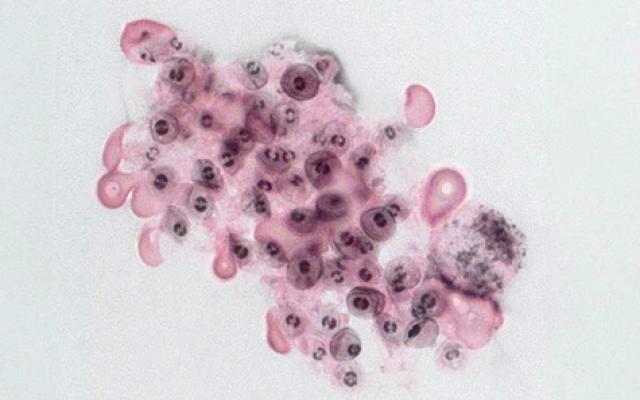
Banner image: Pneumocystis. Credit: Philippe Hauser, CHUV