From SARS-CoV-2 monitoring to HIV antiviral drug resistance detection, the SIB Resource V-pipe orchestrates several software packages to detect the genomic diversity of a virus population in a sample or individual. The pipeline is a key companion for researchers conducting large-scale epidemiological surveillance applications as well as patient-focused clinical applications. On the occasion of the publication of the paper presenting the resource, we asked the team developing it, the Computational Biology Group at ETH Zurich led by Niko Beerenwinkel, to tell us about the tool’s evolution, its recent applications and future developments.
What is V-pipe?
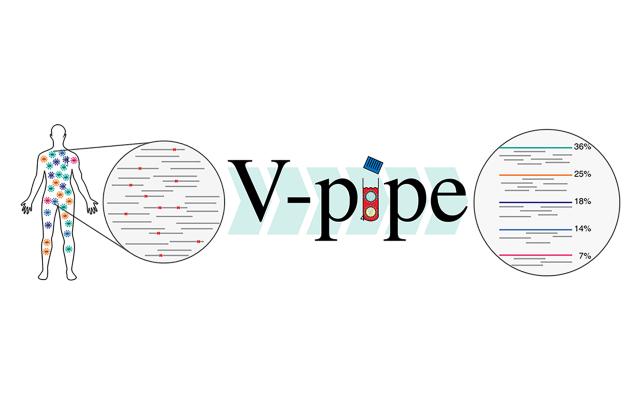
V-pipe is a program for analyzing sequencing data obtained from virus samples in an automated, reproducible, and transparent fashion. It is a bioinformatics pipeline, i.e., an orchestrated collection of computational tools.
How can V-pipe be used in research?
V-pipe can infer the viral genomic diversity present within a sample, based on next-generation sequencing data. Such estimates are important for viral diagnostics and treatment as well as for epidemiological surveillance.
Can you cite a particularly exciting example of a study using V-pipe?
Most recently, we have used V-pipe to detect genetic variants of the coronavirus in wastewater samples. We found several variants of concern, often earlier than in clinical samples. This study, made available as a preprint, has generated interest from researchers worldwide seeking to replicate our approach locally, including from wastewater analysis projects in Austria, Germany, Greece, and indeed, the International Water Association.
On a completely different and unexpected topic, French marine biologists are using V-pipe to detect coinfections of multiple different strains of herpesvirus, which, we learned, is of great concern in oyster farming.
What are the key changes made to V-pipe since its inception?
The pipeline itself has been reengineered in several ways to become more robust and flexible. At the same time, we have improved several of its components, most notably the mutation calling step. For example, these efforts have enabled V-pipe to scale to very large cohorts, such as the thousands of coronavirus samples we have analyzed with V-pipe.
Which feature in V-pipe is currently most exciting?
With the beginning of the COVID-19 crisis we have rapidely provided a SARS-CoV-2-specific branch of V-pipe, which is now used widely and also driving the majority of the Swiss coronavirus surveillance sequencing efforts, including the genomic characterisation of variants of concern.
Thinking about the future: how do you expect to see V-pipe continue to evolve?
We will continue to develop V-pipe for both large-scale epidemiological surveillance applications as well as patient-focused clinical applications. The early detection of emerging mutations in HIV samples, which confer resistance to antiviral drugs, is an example of application of V-pipe to assist medical decisions.
V-pipe joined the portfolio of SIB Resources in 2017 as an emerging resource. What does it mean to be an SIB Resource?
We are benefitting immensely from the support of the SIB, the professional environment of bioinformaticians and software developers, and SIB’s international network.
Reference(s)
Posada-Céspedes S et al., V-pipe: a computational pipeline for assessing viral genetic diversity from high-throughput data. Bioinformatics, 2020.