Scientists at SIB and the University of Basel, and Chelonia Applied Sciences (Basel), are part of a European effort to find molecules active against COVID-19 - and to accelerate this process in the future. They are part of Exscalate4CoV (E4C), a public-private consortium of 18 EU top organizations - including the European supercomputing platform EXSCALATE - from 7 countries, which received 3 million Euros from the European Commission’s Horizon 2020. The scientists will be using the SIB Resource SWISS-MODEL to generate 3D-models of proteins that have not yet been experimentally elucidated, thereby accelerating virtual screening for potential drugs.
Accelerating the search for drugs against COVID-19
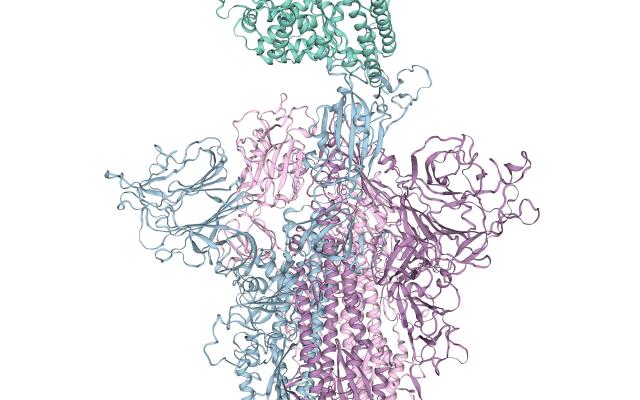
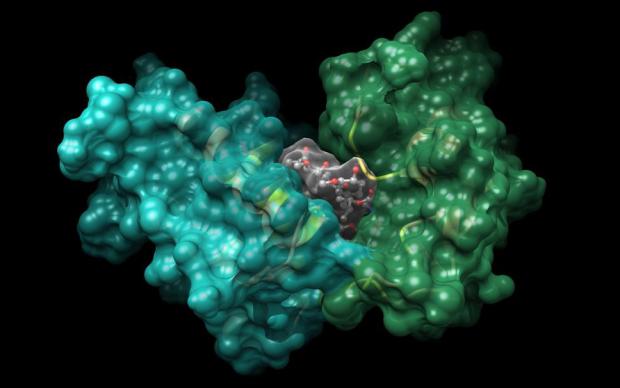
As part of the E4C consortium, the SIB Computational Structural Biology (CSB) Group at the Biozentrum (University of Basel) is in charge of generating predicted three-dimensional models of COVID-19 proteins in silico. “As this is a relatively new virus, scientists only recently started to experimentally characterize the 3D structures of the viral proteins. Computational methods can model structures using information from related proteins, such as from SARS-CoV, thereby accelerating the drug discovery process”, says Gerardo Tauriello, Development Team Leader at CSB. This approach is called homology-modelling, and one of the leading tools to perform it is developed by the team: the SWISS-MODEL server, an open access SIB Resource. Both experimental structures and homology models will be used by the other participants of E4C to identify small molecules potentially capable of targeting the coronavirus. These molecules will then enter further testing in in vitro experiments, animal models and - if successful- clinical trials.
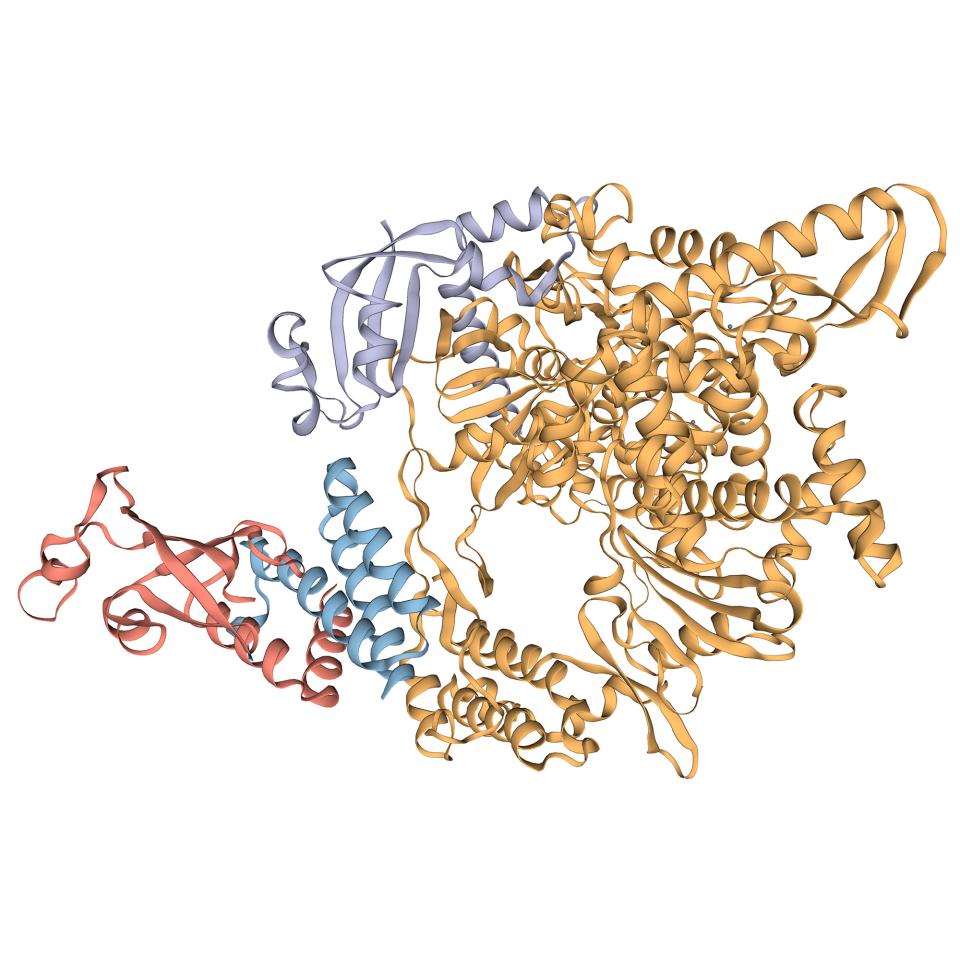
Scrutinizing proteins in 3D to help drug discovery
Viruses, even though they require the machinery of host cells to express their genome, depend on proteins encoded in their genome, in the same way living organisms are. Each protein has a particular three-dimensional structure, tightly linked to its function. For instance, a range of transmembrane proteins are used by the virus to bind to host cells. Drug molecules are designed to bind to relevant parts of a protein in order to modulate its function. A three-dimensional representation of a protein’s structure at an atomic level of detail is therefore a key aspect in rational drug discovery.
Beyond COVID-19: enabling faster drug discovery for future pandemics
E4C aims to leverage the learnings and developments being made during the current outbreak to accelerate the discovery and identification of drugs in the event of future pandemics - in particular when the pathogen is new to science and where no treatment exists. “Today, processing a novel viral genome requires time-consuming manual work by modelling experts which limits reaction time. In the future, as soon as a new virus is sequenced, any researcher will be able to use SWISS-MODEL to generate models within minutes and have them ready to use and interpret within the EXSCALATE platform”, explains Tauriello. As part of E4C, the SIB CSB Group will develop and benchmark an automated and scalable workflow to enable the generation of protein models by homology, directly from the genome of any virus.
Coupling top life-science research labs with powerful supercomputing
E4C is coupling the European supercomputing platform EXSCALATE (EXaSCale smArt pLatform Against paThogEns) - able to leverage a library of 500 billion drug candidates, thanks to a processing capacity of more than 3 million molecules per second - with top life-science research labs. Chelonia Applied Science, based in Basel, designed the E4C consortium, and is in charge of its dissemination and exploitation activities. Silvano Coletti, Chelonia’s Chief Executive Officer, says: “The situation the world is facing with the COVID-19 pandemic is unprecedented in our lifetime, impacting us all professionally and personally. Exscalate4CoV is an equally unprecedented computational effort, which aims to accelerate COVID-19 drug identification today, and to become the reference platform for future pandemics tomorrow - including discovery and development process in pharmaceutical research.”
Additionally to the Swiss members, the E4C consortium, coordinated by the Italian Pharma Dompé, includes 16 institutions* from six countries. Learn more on the EXSCALATE4CoV website.
*Dompé Farmaceutici SPA, Politecnico di Milano (Dept. of Electronics, Information and Bioengineering), Consorzio Interuniversitario CINECA (Supercomputing Innovation and Applications), Università degli Studi di Milano (Pharmaceutical science Department), Katholieke Universiteit Leuven, International Institute Of Molecular And Cell Biology In Warsaw (LIMCB), Elettra Sincrotrone Trieste, Fraunhofer Institute for Molecular Biology and Applied Ecology, Bsc Supercomputing Centre, Forschungszentrum Jülich, Università Federico II di Napoli, Università degli Studi di Cagliari, KTH Royal Institute of Technology (Department of Applied Physics), Associazione BigData, Istituto Nazionale di Fisica Nucleare (INFN) and Istituto nazionale per le malattie infettive Lazzaro Spallanzani.